import pandas as pd
import numpy as np
from plotnine import ggplot, aes, after_stat, geom_bar, labs
Evaluate mapping after statistic has been calculated
Parameters
x : str-
An expression
See Also
Examples
after_stat
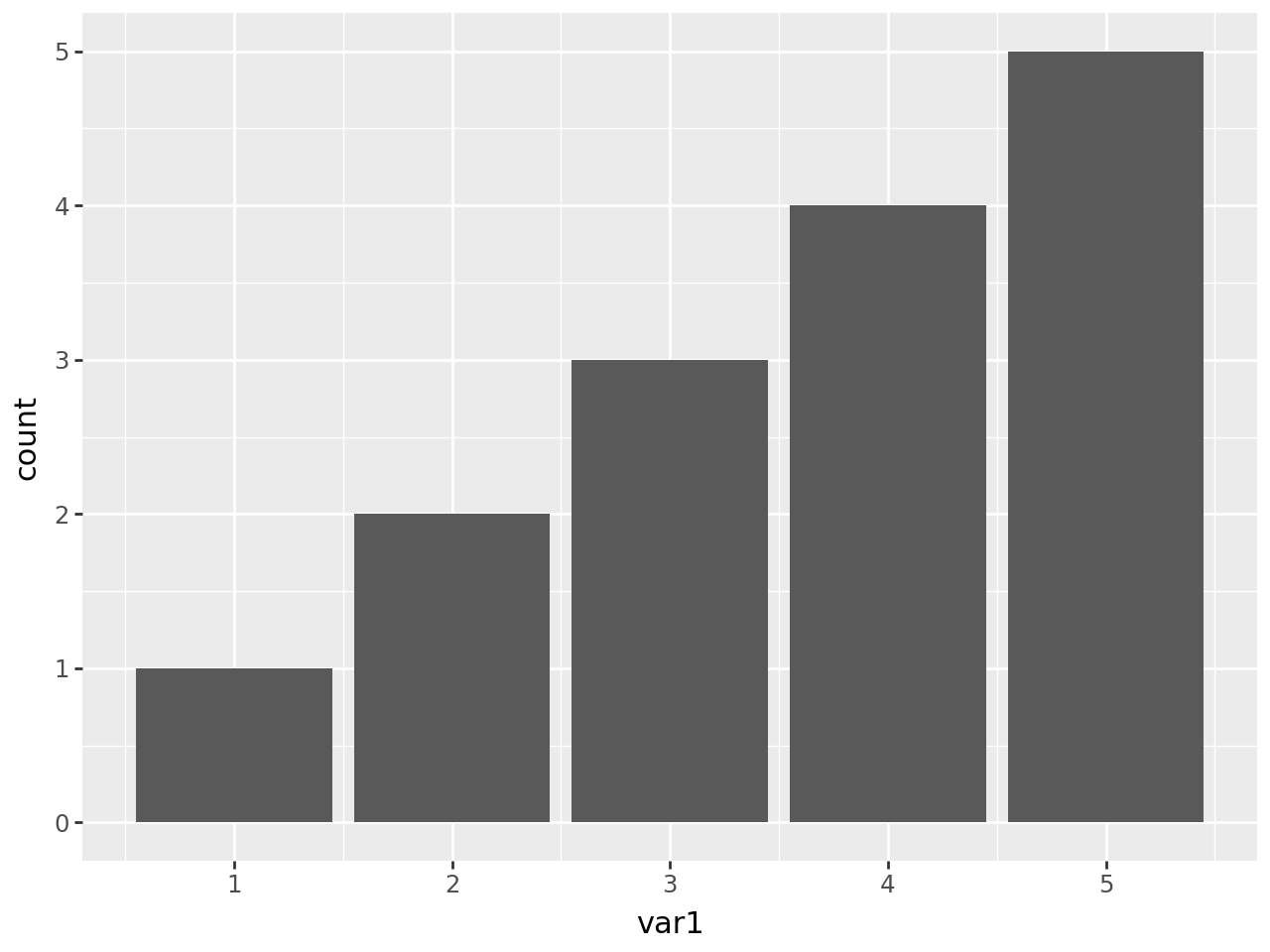
geom_bar uses stat_count which by default maps the y aesthetic to the count which is the number of observations at a position.
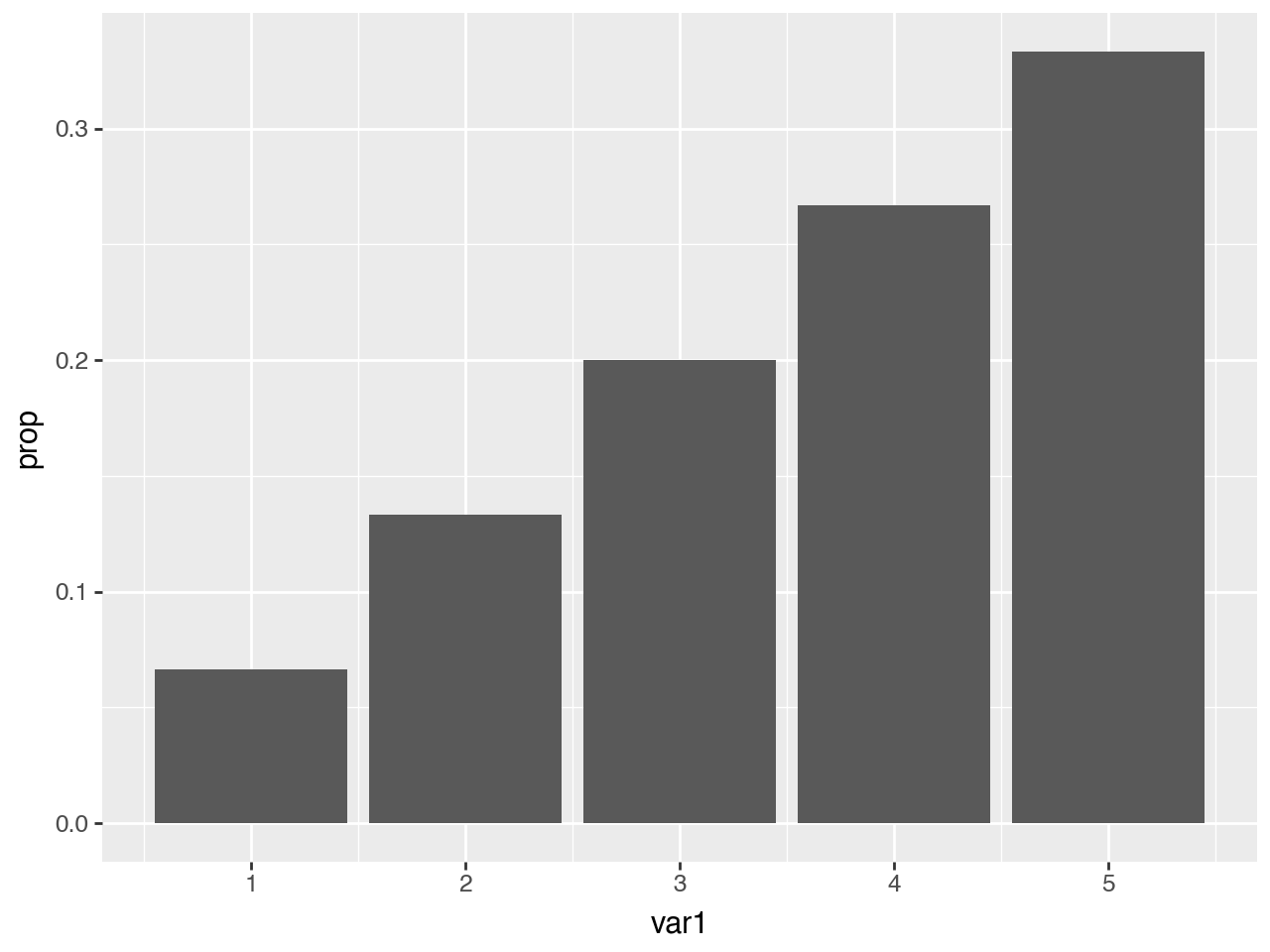
Using the after_stat function, we can instead map to the prop which is the ratio of points in the panel at a position.
With after_stat you can used the variables calculated by the stat in expressions. For example we use the count to calculated the prop.
By default geom_bar uses stat_count to compute a frequency table with the x aesthetic as the key column. As a result, any mapping (other than the x aesthetic is lost) to a continuous variable is lost (if you have a classroom and you compute a frequency table of the gender, you lose any other information like height of students).
For example, below fill='var1' has no effect, but the var1 variable has not been lost it has been turned into x aesthetic.
We use after_stat to map fill to the x aesthetic after it has been computed.